Abstract: Post-translational modification(PTM) plays an important role in regulating protein function. Currently, over 600 modifications have been found in mammalian cell protein. Besides typical PTMs like phosphorylation, glycosylation, acetylation, ubiquitination etc, more and more new modifications are found(e.g. succinylation). Applications in human cancer treatment and prevention remain to be further explored.
Keywords: Post-translational Modification, Phosphorylation, Glycosylation, Ubiquitination, Acetylation, Methylation, ADP-ribosylation
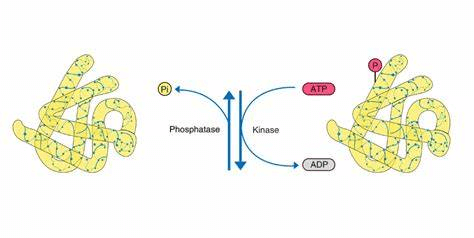
1. Phosphorylation
Phosphorylation modification is reversible and usually occurs at serine, threonine or tyrosine residue. It's one of the most studied PTMs. 30% proteins in mammalian cell are phosphorylated at one or more sites. Improper protein phosphorylation causes many human diseases.
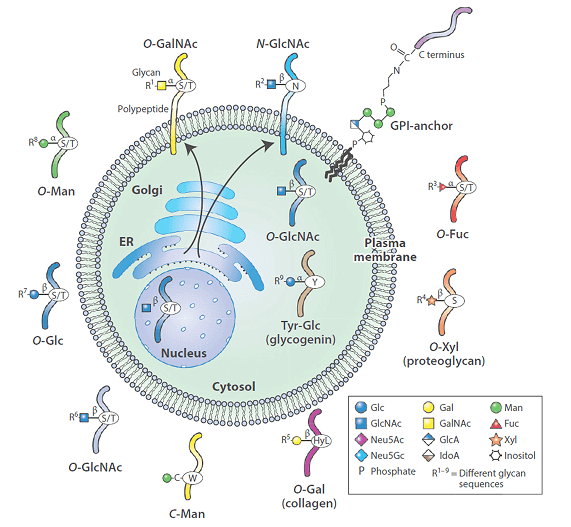
2. Glycosylation
Glycosylation widely exists in various biological environment and has a great effect on folding, conformation, distribution, stability and activity of protein.
There are two types of mammalian glycosylation. The carbohydrate which exists in the form of oligosaccharide linked by aspartate(N-linked) or serine/threonine(O-linked) is the main structural component of many cell surface and secretory proteins. Protein glycosylation can change the conformation, oligomerization and conversion of protein, resulting in the change of cell signaling pathway.
The glycosylation of cell membrane and secretory proteins is shown in the figure below.
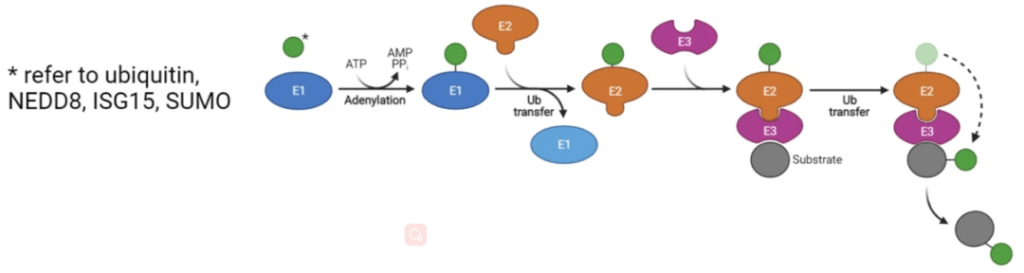
3. Ubiquitination
Ubiquitin(Ub) is a kind of polypeptide(MW: 8 kDa). Through enzyme cascade among ubiquitin-activating enzyme(e1), ubiquitin-conjugating enzyme(e2) and ubiquitin ligase(e3), Ub molecule adheres to substrate protein lysine residue to produce monoubiquitination, polyubiquitination and branched ubiquitination. This is adjusted by deubiquitinase. Besides ubiquitination, there are some ubiquitin-like molecules, including SUMO, NEDD8, ISG15, FAT10, ATGs, HUB1, FUB1. They bind with their substrates through the ubiquitin-like enzyme linked reaction.
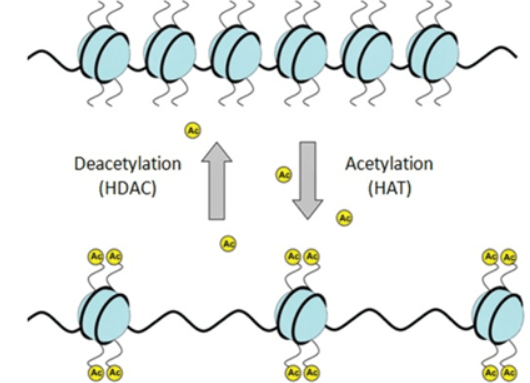
4. Acetylation
Catalyzed by acetyltransferase or non-enzyme, acetyl group is transferred and added into lysine residue or N-terminal.
There are two types of acetylation modification: N-terminal and lysine acetylation. 80-90% eukaryotic protein is N-terminal acetylation.
Early acetylation research mainly focuses on histone in the nucleus and regulation of transcription and gene expression. Many transcription coactivators have the intrinsic activity of acetyltransferase. Transcription co-repressors are related to the activity of deacetylase. With the development of mass spectrometry, many non-histone in cytoplasm or other organelles are modified by lysine acetylation. The acetylation of special sites of more and more non-histone can regulate the activity, localization, specific interaction, stability and degradation.
Acetylation is a common and important protein PTM, which has a great effect on chromosomal structure of cells, activation of transcriptional regulators in nuclei, cell cycle and metabolism, actin polymerization control, and tumors and polyglutamine diseases etc.
Protein acetylation is also the important target for new drug development and design in recent years.
5. Methylation
Protein methylation usually occurs at lysine or arginine residue. Lysine can be mono-, di-, or tri-methylated by lysine methyltransferases(kmt) and then removed by lysine demethylases(kdm). Arginine can be monomethylated by PRMTs, dimethylated symmetrically or asymmetrically.
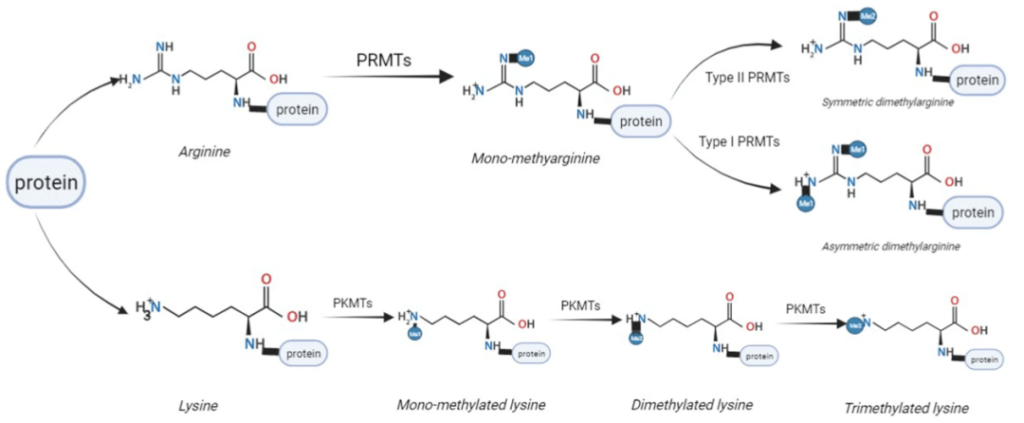
Protein methylation and acetylation are common epigenetic modification. They usually occur at histone. Besides histone, methylation can occur at many proteins. Protein methylation may affect protein-protein interaction, protein-DNA interaction, or protein-RNA interaction, protein stability, subcellular localization or enzymatic activity. The methylation modification of many transcription factors can affect gene expression.
6. ADP-ribosylation
ADP-ribosylation is a highly conserved PTM with one or more ADP riboses added on the target protein, including mono-ADP-ribosylation(MARylation) or poly-ADP-ribosylation(PARylation). ADP-ribosylation is a kind of dynamically reversible PTM system, regulated by enzymes like writer, reader, eraser. Writers can catalyze the formation of ADP-ribosylation modification and are called ADP-ribosyl transferase(ADPRT, ART) which is a kind of protein with mono- or poly- ADP-ribosyl transfer activity.
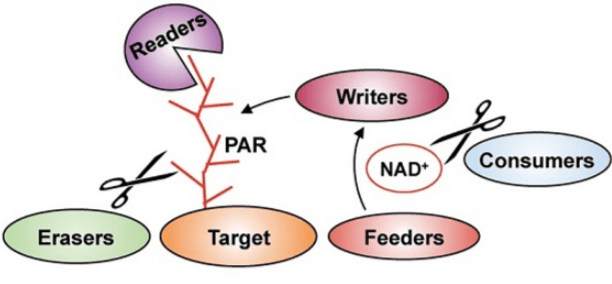
ADP-ribosylation can change the activity, stability, ligand binding ability of proteins and has a great effect on cell function. Bacteria prevent virus infection and antagonize host immune system by MARylation. They also attack key molecules in eukaryotic host cells, resulting in the disastrous consequences in the cell.
7. Recommended Recombinant Proteins
- 【P1746】Recombinant Human CTBP2
- 【P1238】Recombinant Human SESN2
- 【P3003】Recombinant SARS-CoV-2 Nucleoprotein
- 【P3006】Recombinant SARS-CoV-2 Spike Glycoprotein RBD
- 【P0010】Recombinant Human PCT
REFERENCES
[1]Post-translational modification enzymes as key regulators of ciliary protein trafficking, PMID: 33681987.
[2]Decoding Post-Translational Modification Crosstalk With Proteomics, PMID: 34339852.
