Abstract: Prokaryotic expression refers to the method of inducing exogenous gene into expression strain by establishing expression vectors and making it expressed in prokaryotes or cells with gene cloning technology. Currently, prokaryotic protein expression system is the most cost-effective and commonly used protein expression method. E. coli. expression system is one of the widest applied prokaryotic protein expression systems. A complete expression system usually includes matching expression vectors and expression strains. Special inducible expression also includes inducer. Fusion expression includes purification system or tag detection etc. Selection of expression system usually depends on experimental purpose, such as low/high expression level , activity of target protein, purification methods of expressed products etc. Common questions during prokaryotic expression process are introduced below.
Keywords: Protein Synthesis in Prokaryotes, Prokaryotic Expression, Protein Expression
1. Why does the target protein always appear in the form of inclusion body?
In prokaryotic expression and purification, the target protein is often mistakenly folded and aggregates to form inclusion body. The expression of induced target protein usually can reach over 50% of total cell protein. Although a certain percentage of the protein can exist in the form of soluble form, over 95% protein exists in inclusion body.
During the experimental process, more soluble proteins can be obtained by decreasing induced temperature (e.g. 16-30℃), or IPTG concentration (0.01-0.1mM) with extended induced time. Using special medium to culture bacteria can get more soluble proteins as well.
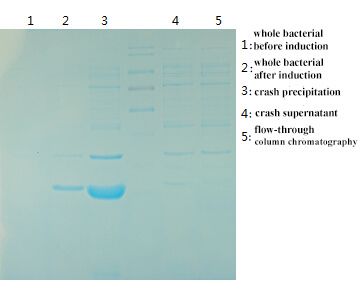
2. Why is the transmembrane protein hard to express?
The low expression success rate of transmembrane protein is a common question. There are two main reasons.
2.1. Transmembrane Chemical Structure
The transmembrane protein usually is the simplest transmembrane chemical structure formed by the saltatory binding of hydrophobic amino acid molecule and hydrophilic molecule. Its structure is similar to the structure of signal peptide. It is difficult for for prokaryotic cells to complete the complex process including recognition and cutting of signal peptide, induction of endoplasmic reticulum, repackaging and secretion of golgi apparatus like eukaryotic cell. Some proteins are transmembrane many times. It is hardly possible for prokaryotic cells to complete the mission.
2.2. Hydrophobic Fragments
It's very easy for hydrophobic fragments to form inclusion body in prokaryotic expression. Hydrophobic polypeptide inhibits the translation process and even fuses with prokaryotic membrane structure to form toxicity. Since organisms have the instinct for self-preservation, all organelles will stop the process for synthesizing protein.
3. How to choose host bacteria for protein expression?
Codons preferred by prokaryotic expression system and eukaryotic cell are different. Hence, when eukaryotic genes are expressed in prokaryotic system, some codons in eukaryotic genes may be rare codons for prokaryotic cells. As a result, the efficiency and level of expression are very low. Problems for protein expression and strain selection are outlined below:
3.1. There is no protein expression or protein truncation happens
Possible reasons are codons deviation of escherichia coli. Rare codon tRNA shall be added. Alternatively rare codons are mutated into common codons of escherichia coli.
Recommended expression strains: Rosetta 2, Rosetta-gami 2, Rosetta-gami B, RosettaBlue
3.2. Expressed as inclusion body protein
Two possible reasons: I. target protein contains disulfide bond which is hard to form. Vector tags can be used to advance the formation of disulfide bond. Recommended expression strains are Origami 2, Rosetta-gami 2 and Rosetta-gami B; II. The expression is too fast and the expression level is too high. The decrease of induced temperature and optimization of IPTG concentration can solve the problem. Recommended expression strains are Tuner and Rosetta-gami B.
3.3. The protein is inactive
The protein may be mistakenly folded. Various vector tags for promoting the formation of disulfide bond can be used. Recommended expression strains are Origami 2, Rosetta-gami 2 and Rosetta-gami B. Decreasing induced temperature and optimizing IPTG concentration are also applicable. Recommended expression strains are Tuner and Rosetta-gami B.
3.4. The cell is dead or has a great difficulty in its growth
The expressed protein may be toxic. Background expression needs to be more strictly controlled. Recommended expression strain is pLysS.
4. Prokaryotic protein purification method and selection
Common protein purification methods are affinity purification, ion exchange and gel extraction.
4.1. Proteins with fusion tags
Proteins with fusion tags like HIS,GST,SUMO,Fc etc can be affinity purified by Ni column, GST column and protein A. It is convenient and fast to use affinity purification method to get proteins with over 85% purity.
4.2. Proteins without tags
If the recombinant protein is not expected to contain tags, recombinant protein with tags can be expressed first. After affinity purification, tool enzyme can be used to cut and fuse the protein. After repurification, the tool enzyme shall be removed. Alliteratively, recombinant proteins without tags can be directly expressed. Purification processes including ion filtration, gel filtration and hydrophobic interaction can be performed.
4.3. Expressed protein is used as antigen
If the expressed protein is used as antigen, the gel extraction can be directly conducted to obtain proteins with higher purity. The sensitivity of immunized antibody used for WB is higher.
| Characteristics | Techniques |
| Size | Gel filtration(GF), also called molecule screening |
| Charge | Ion Exchange Chromatography(IEX) |
| Hydrophobicity | Hydrophobic Interaction Chromatography(HIC) |
| Reversed Phase Chromatography(RPC) | |
| Bioidentification(Ligand Specificity) | Affinity Chromatography(AC) |
5. Is the obtained protein active?
If the recombinant protein expressed by prokaryotic system is expected to be active, conditions are very complex. Differences of protein activity are influenced by suitable buffer solutions, salt concentration, protein folding state and even the difference of method for detection activity. Thus, we try to get the protein from supernatant and expect protein folding to be close to the active state.
6. FineTest Recombinant Protein
Browse a list of FineTest recombinant protein and contact us to order your protein products.
